Hello VirtualFlow Community!
I’ve sucesfully finished a test run,but during the post processing I am meeting some problems.
The command:
head -n 100 …/…/ranking/qvina02_rigid_receptor1/firstposes.all.minindex.sorted.clean > compounds
creates a list called compounds containing the top 100 docking poses with their respective score.
However, after running
vfvs_pp_prepare_dockingposes.sh …/…/…/output-files/complete/qvina02_rigid_receptor1/results/ meta_collection compounds dockingsposes overwrite
problem occurred:
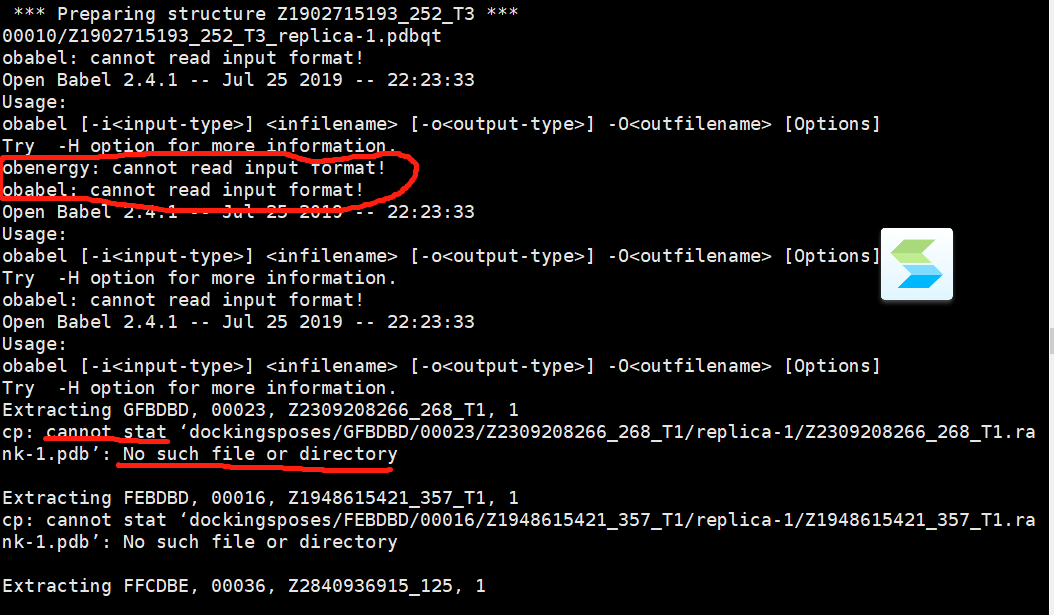
get two folders and three files, like this

but the dockingsposes.plain is empty. How to alter the command or fix the problem? And how to prepare the input ligands file using this dockingsposes result files to run next flexible docking?
Could you help me?
Sincerely,
Vera
