Hi,
I am trying to run VFVS and I am encountering the following problem. Everything seems to work fine, but the jobs finish before all ligands are docked. When looking at the output files, a few ligands per collection are docked and the rest just seems to be skipped. I have tried different ligand libraries and also different receptors and can’t find a pattern in which ligands are skipped. When running the same workflow multiple times, the results are not consistent. I also can’t find any error messages.
Apparently JanetZ had a similar problem in September 2020 (Why there were just part of my ligands been docked?, but I couldn’t find a solution under the topic.
I have attached the report of the “completed” workflow, any help or tips would be greatly appreciated.
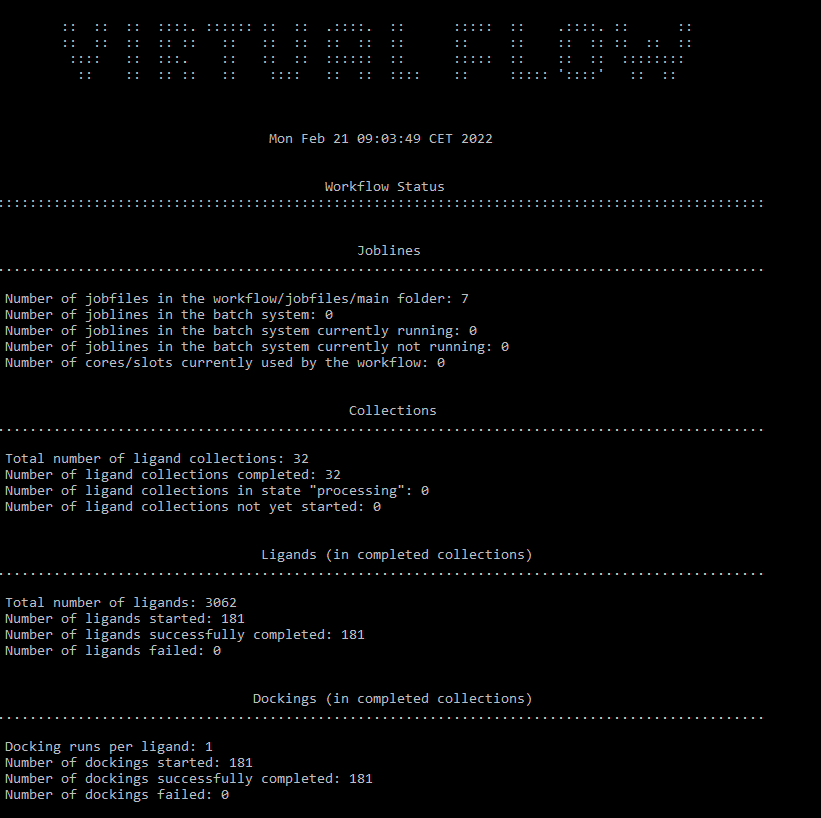
Using the vf_continue_all.sh script should continue the job lines where they left off.
If you have further issues, you may be able to diagnose them by looking at the error log files. If I remember correctly, they are located in <temp_directory>/<<job_letter>/. You should be able to find some log files with “err” in the file name and they may give some hint as to why the job lines are failing.
Hi Alan,
thank you so much for your reply.
I’ve tried using vf_continue_all. and it just starts the jobs and then finishes them right away. The jobs don’t fail or abort in the first place, they just finish without having run all ligands. It starts docking a couple of ligands of one collection, then say the collection is completed and go to the next, so it does “complete” the jobs in a sense.
I also can’t find error logs for the jobs and the error logs for the queues are empty.
I ran the preconfigured tutorial and it works fine, so I’m assuming the problem is either in my setting or, most likely, in my ligand-library, since VFLP didn’t work for me either. Do you have any advice what might cause such a problem or what to watch out for?
Can you please post the results of cat all.ctrl|grep -v "#"|sed '/^$/d'. What is your lib size in terms of total number of ligands? How many collections these ligands are distributed in? I had a similar issue before, which went away after I set the ligands_todo_per_queue and the ligands_per_refilling_step parameters correctly. As not many ligands are docked yet, it may be a good idea to start fresh with the correct parameters.
Hi, thank you for your advice.
I have tried different things, so I will just post one example of the output you asked for:
*****************************************************************************************************************************************************************
*************************************************************** Job Resource Configuration ****************************************************************
*****************************************************************************************************************************************************************
job_letter=r
batchsystem=SLURM
partition=medium
timelimit=1-00:00:00
steps_per_job=1
cpus_per_step=8
queues_per_step=8
cpus_per_queue=1
*****************************************************************************************************************************************************************
********************************************************************* Workflow Options ********************************************************************
*****************************************************************************************************************************************************************
central_todo_list_splitting_size=600
ligands_todo_per_queue=100
ligands_per_refilling_step=20
collection_folder=../input-files/ligand-library
ligand_library_format=pdbqt
minimum_time_remaining=10
dispersion_time_min=3
dispersion_time_max=10
verbosity_commands=standard
verbosity_logfiles=standard
store_queue_log_files=all_compressed_error_uncompressed
keep_ligand_summary_logs=true
error_sensitivity=normal
error_response=fail
tempdir_default=/tmp
tempdir_fast=/dev/shm
outputfiles_level=collection
prepare_queue_todolists=true
*****************************************************************************************************************************************************************
***************************************************************** Virtual Screenign Options ***************************************************************
*****************************************************************************************************************************************************************
docking_scenario_names=qvina_w_rigid_receptor1
docking_scenario_programs=qvina_w
docking_scenario_replicas=1
docking_scenario_inputfolders=../input-files/qvina_w_rigid_receptor1
*****************************************************************************************************************************************************************
******************************************************************* Terminating Variables *****************************************************************
*****************************************************************************************************************************************************************
stop_after_next_check_interval=false
ligand_check_interval=10
stop_after_collection=false
stop_after_job=false
At the moment I have just over 3000 ligands distributed in collections of just below 100 each, so 31 collections. I have, however, tried different distributions. If I get it to work, I also plan on using a larger library (about 10x as many). I have also tried adjusting the two settings you mentioned, but I can’t seem to get it right. Is there a rule or guideline on how to set those? Thanks in advance
Yes there is some rules for the settings- I think I found it in the forum somewhere, can not recall. The documentation is nowhere complete. What happens when you set the following parameters:
cpus_per_queue=8 # I used the recommended setting as per the comments in all.ctrl
central_todo_list_splitting_size=1000 # the recommended setting is this number should be high, becomes an issue when the lib size is very big.
ligands_todo_per_queue=400 # if each queue does this number the lib should be complete
ligands_per_refilling_step=100 # average number of ligands per collection
Start afresh, i.e. do not use vf-continue.all.sh
Thank you for the advice, I used the settings you suggested, but ran into the same issue.
However, I now got it to work. Apparently there were some issues with the ligand library and having previously opened it in Windows. I prepared a new one in Ubuntu and now it works fine.