Dear VirtualFlow community,
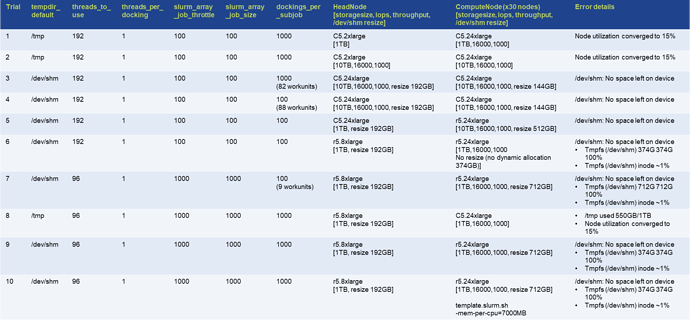
In VirtualFlow, I think the compounds (input-files/ligand-library) used for docking are once copied and expanded to the directory specified by tempdir_default.
Even if the capacity of /dev/shm is expanded up to 712GB in the case of more than 10 million compounds, it becomes "No space left on device" and all ligands cannot be deployed.
So we changed tempdir_default to /tmp and reserved 1TB of capacity, and only about 550GB is used (achieving a write speed of 1GB/sed) and CPU utilization of the each node is only about 15%.
Could you please advise on specific variable settings (e.g.parameters in all.ctrl and storage size for /dev/shm) that will realize high performance when dealing with 10 million+ ligands. Thank you in advance. We believe VirtualFlow is an innovative tool in drug discovery.
Attached are the results of the study to date in AWS ParallelCluster using Slurm.