When I prepare the docking poses using vfvs_pp_prepare_dockingposes.sh, I get an error about a file that says cannot open ‘compound.energies’ for reading, however there is a very similar file (I’ve encircled it). However in the script vfvs_pp_prepare_dockingposes.sh, it seems as it is looking for a file with “.energies” only. Any idea on what I can do?
Edit: Based on the script vfvs_pp_prepare_dockingposes.sh it seems like compounds.energies.uniq.csv is supposed to contain output based on compound.energies. Also the file firstposes.all.minindex.sorted.clean is empty for some reason.

Hi,
I have still not been able to find a solution to this problem. It’s frustrating since I got the job to run, and the scripts that have been provided don’t seem to be providing the right output for me. Any advice?
Dear Janujan,
Welcome to the forum.
In the meantime, I have added a new script which does everything automatically:
- Creation of a complete ranking
- Creation of DataWarrior files
- Preparation of the docking poses
The script has the name “vfvs_pp_all.sh”, and needs to be run in the VirtualFlow root folder. See also the help command of the script. This script deploys the other scripts (including the ones you used) automatically.
Does this script work for you?
Best,
Christoph
1 Like
Thanks Christoph for the response. I have yet to run the script “vfvs_pp_all.sh” as the connection to my cluster is down right now, but if your script “vfvs_pp_all.sh” executes the same script that is giving me the errors, won’t I get the same error? Once the connection comes back on, I will run the script and let you know how it goes anyways.
If you used all the scripts correctly, it should have worked. It is easy to pass an incorrect argument to one of the scripts for example. This problem is avoided with the new wrapper script.
If the wrapper script also fails, then there might be another problem. Let’s see what the result with vfvs_pp_all.sh is. Please make sure you have the VFTools/bin folder in your PATH variable.
Good luck 
1 Like
Hi Cristoph,
Thanks once again for the prompt responses. When using “vfvs_pp_all.sh”, where is the path for <smiles_collection_folder> option? The output format was pdbqt rather than SMILES for me (if that matters).
Also, what is a “dwar” compound? Not sure what to specify for that parameter.
Cheers
Hi Janujan,
Ok, now I remember why I did not yet publicly announce the new script. For that, you need a few additional ingredients/files.
Let’s try it at first again in the way you did it.
On the cluster, when you run the command vfvs_pp_prepare_dockingposes.sh , is the command obabel available?
Best,
Christoph
Yes it is. I checked using ‘man obabel’. Also turns out the relative path on the VirtualFlow website in the command is wrong, it is supposed to be “…/…/output-files” rather than “…/…/…/output-files”. However that doesn’t fix my error unfortunately.
Could you check if the file “compounds” actually contains some content?

It is empty. I ran the command “head -n 100 …/ranking/vina_files/firstposes.all.minindex.sorted.clean > compounds” prior to using “vfvs_pp_prepare_dockingposes.sh”, so this is strange. Any idea why this might be?
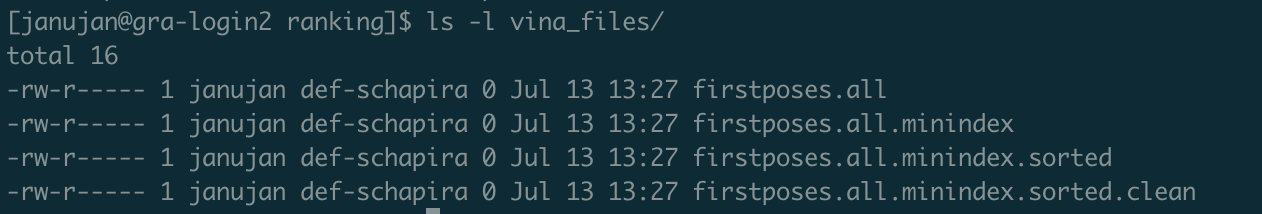
Seems like “firstposes.all.minindex.sorted.clean” is empty as well. Where is “firstposes.all.minindex.sorted.clean” getting populated?
Apologize for spamming the replies. I got it to work. Turns out I had to run the command “vfvs_pp_ranking_all.sh …/…/output-files/complete/ 1 meta_collection”. I had meta_tranche instead of meta_collection. All the files are populated correctly now. I greatly appreciate your help Cristoph!
1 Like
Great, glad you got it to work 
1 Like




